Mills Lab Research Overview
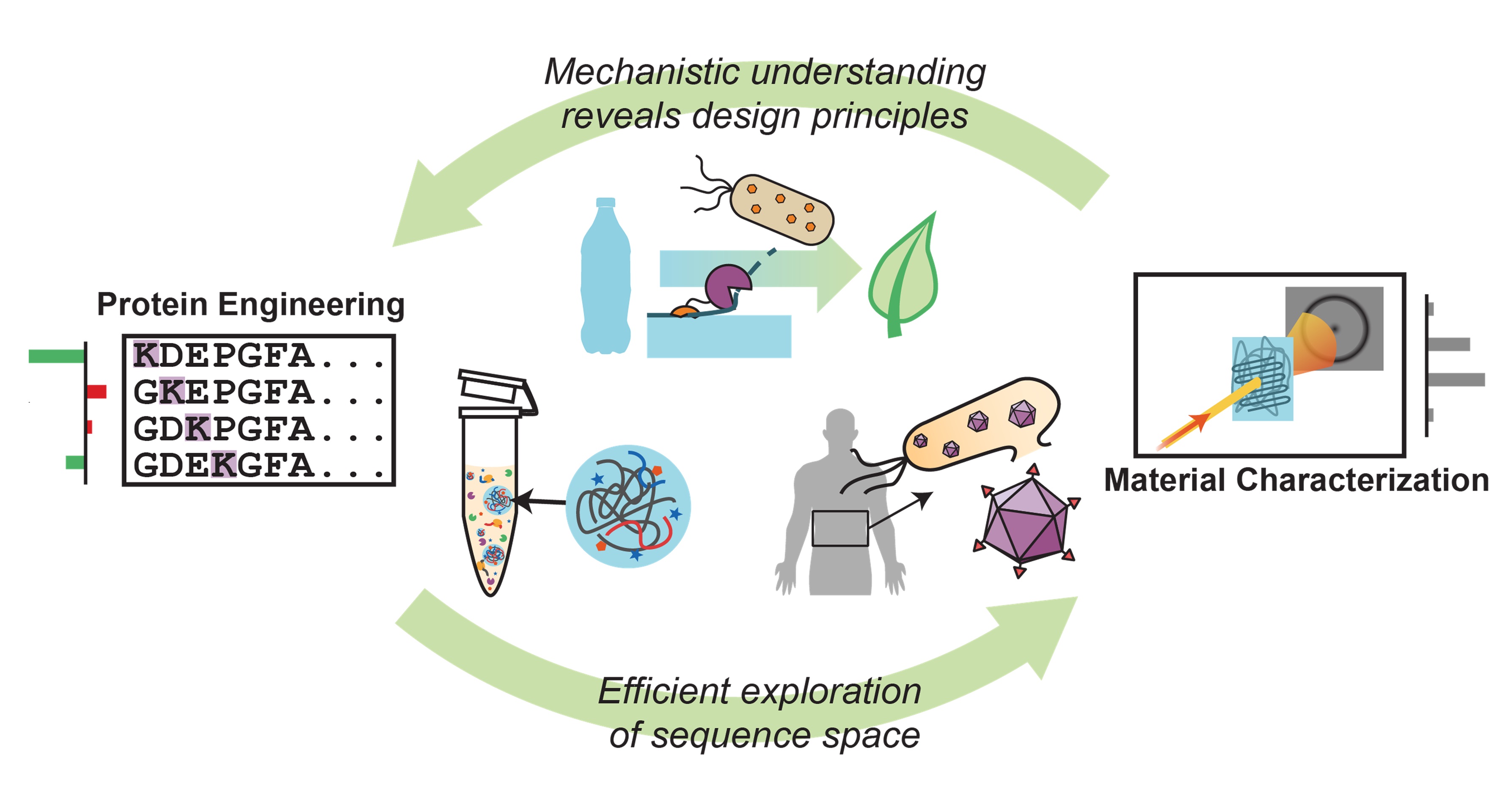
The Mills Lab applies a combination of approaches from polymer science, protein engineering, and synthetic biology to manipulate and understand protein-driven spatial organization in biological systems. Characterization techniques from soft matter provide molecular-level insights into mechanisms of phase separation and protein organization, while protein engineering and synthetic biology techniques enable rapid generation of new protein designs.
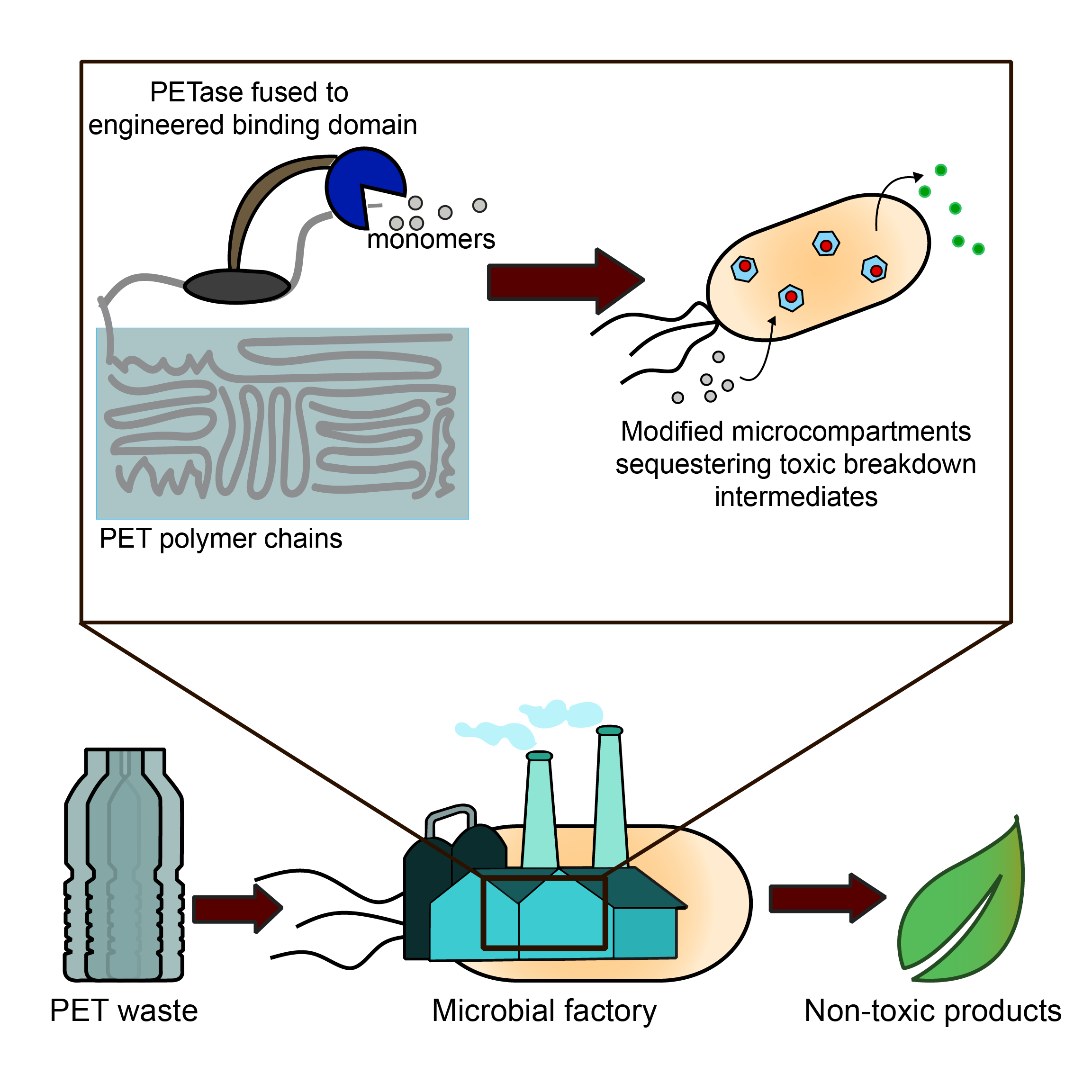
Engineering Spatial Organization Towards Decentralized PET Waste Remediation
Plastic pollution has produced over 200 documented instances of negative impacts on marine life and costs an estimated $13M to the fishing, tourism, and shipping industries. To address plastic contamination in the environment, our lab aims to develop a closed-loop plastic degradation system that is self-contained within engineered microbes. We are applying protein engineering principles to develop tools that enable closed-loop plastic degradation by microbial chasses. Specifically, we are employing spatial organization strategies to improve enzymatic plastic degradation under mild conditions and to convert degradation products into non-toxic molecules.
Developing Cell-Free Systems for Characterizing Protein Liquid-Liquid Phase Separation
Protein liquid-liquid phase separation (LLPS) continues to gain recognition as an essential control mechanism in biology, from gene regulation to viral invasion and proliferation. Cell-free systems provide a bridge between in vivo and in vitro systems that allow a high degree of system manipulation without the loss of biological complexity. We use these systems to study protein liquid-liquid phase separation so that we can engineer it and understand the underlying driving forces for it.
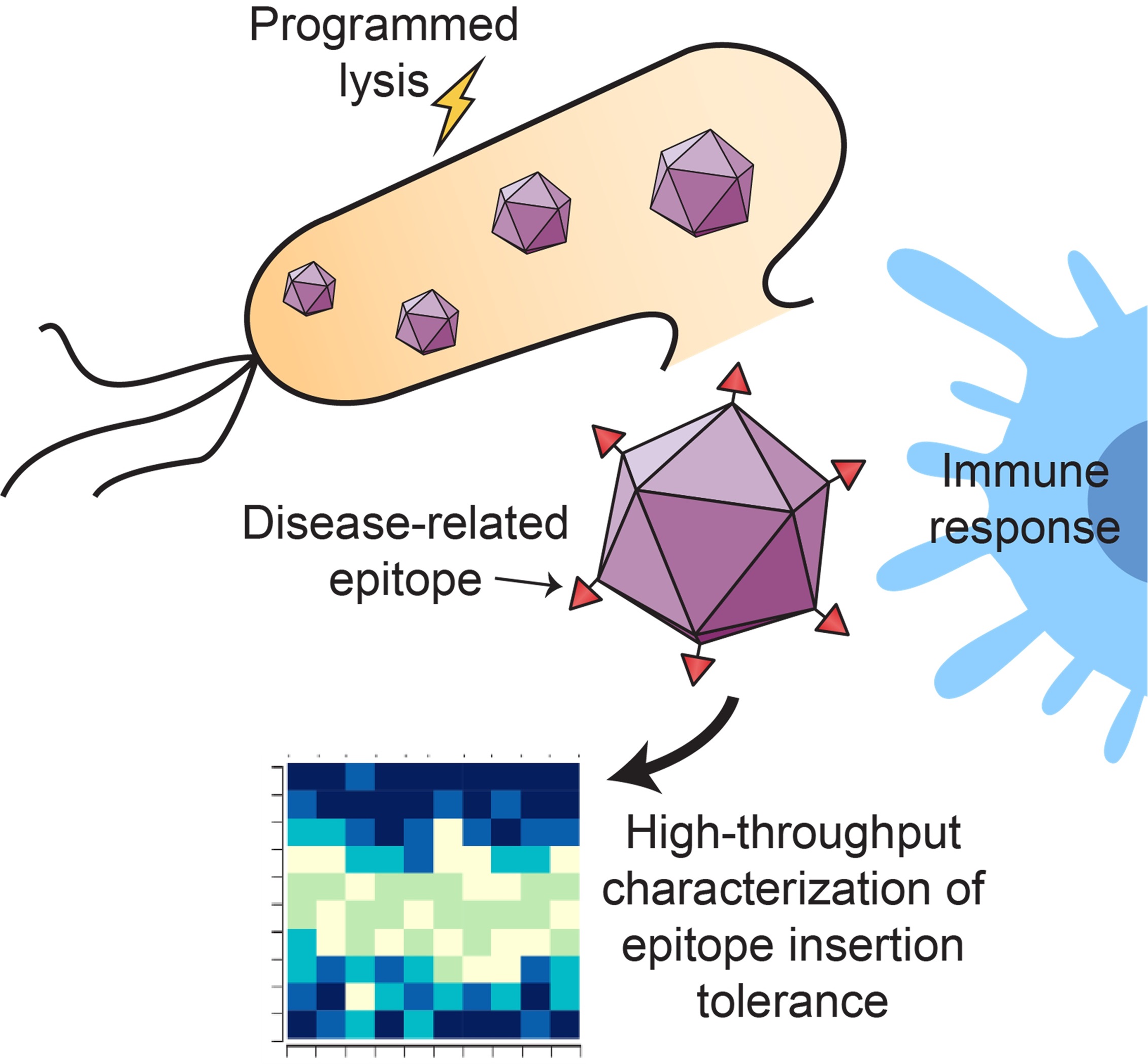
Low-cost Oral Vaccine Delivery with Probiotic Vehicles
Oral vaccines have several advantages over parenteral vaccines, especially in resource-limited settings—they require minimal staffing for administration, circumvent issues associated with needle use, and stimulate both mucosal and systemic immunity. Of oral vaccine candidates, virus-like particles (VLPs) are an attractive option. VLPs are nanostructured protein assemblies made up of viral coat proteins, but without any of the viral infection machinery. Our lab is combining protein particle engineering with programmed lysis circuits for the rapid development and low-cost deployment of these promising vaccine scaffolds.
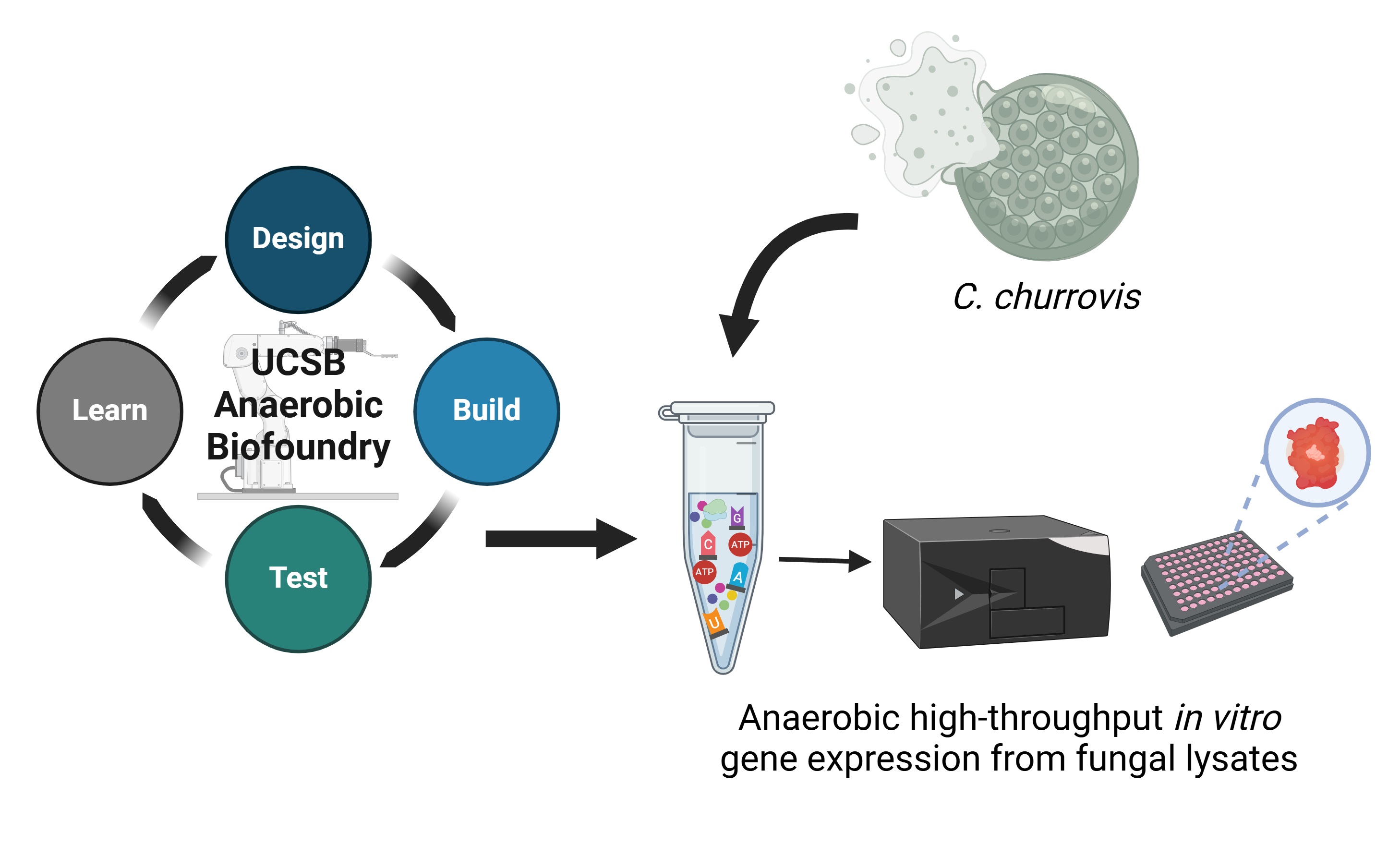
Developing an anaerobic cell-free gene expression system for the screening and rapid prototyping
of anaerobic gut fungal pathways
In collaboration with Prof. Michelle O'Malley and ExFAB (UCSB's NSF Biofoundry)
Anaerobic microbes have evolved unusual abilities that permit them to thrive in the most extreme environments on earth. However, they are often recalcitrant to culture and very difficult to genetically modify, hindering hypothesis testing about protein and pathway function. As a result, nearly 75% of protein encoding genes in anaerobic fungi lack putative function. To address this limitation, our lab is engineering synthetic cell systems inspired by anaerobic fungi and traditional cell-free protein synthesis platforms. We will use these synthetic cell systems to prototype enzymes and pathways that reveal new mechanisms of anaerobic lignin breakdown.